Intel And UPenn Pioneer Machine Learning That Dramatically Improves Brain Tumor Detection
The research specifically took aim at glioblastoma, which the researchers say is the most common and fatal adult brain tumor. Despite expanded treatment options in recent decades, overall survival rates have not significantly improved. Median survival time is around 14 months with standard treatment, or just 4 months without.
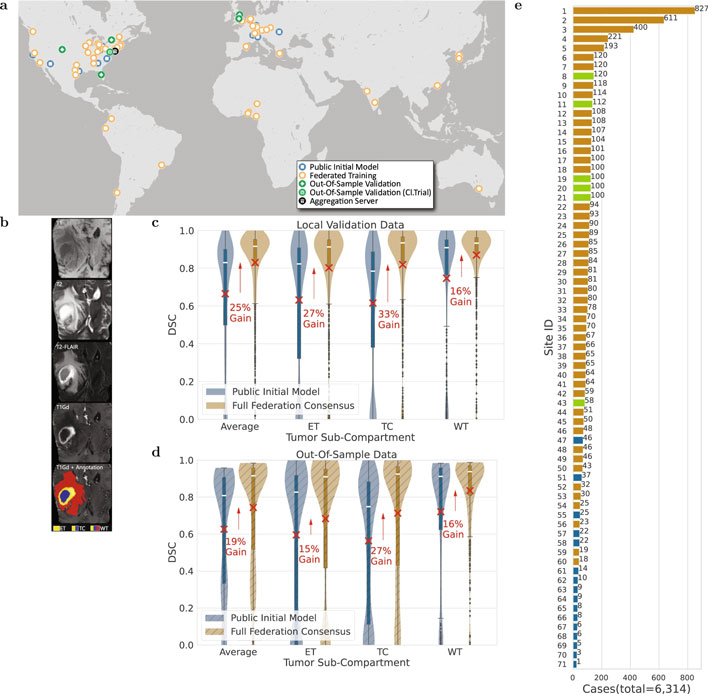
One of the biggest challenges to applying machine learning to medical research is the process of centralizing data while preserving patient privacy. In contrast to centralized learning (CL), where all data is aggregated into a single bucket, FL models allow each partner to retain its data locally. FL models are able to span multiple sites and organizations by sharing only numerical model updates, thus complying with HIPAA and other healthcare regulatory requirements. In this study, data was able to be collected from 71 sites across 6 continents. This kind of global scope is invaluable when studying rare conditions like a glioblastoma, which currently afflict about 3 in 100,000 individuals.
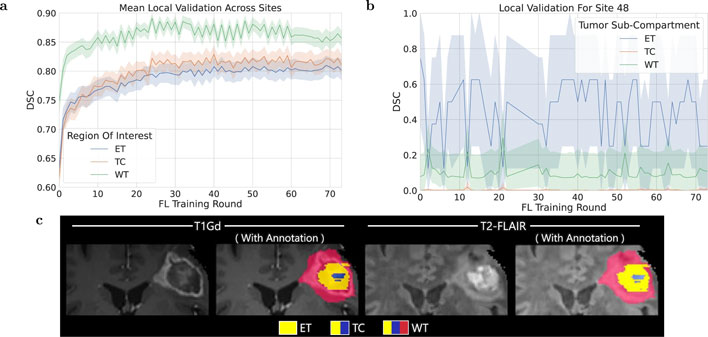
The learning process itself was applied to multi-parametric magnetic resonance imaging (mpMRI) scans which can reveal the presence of the glioblastoma in the brain. These tumors consist of three main sub-compartments, and distinguishing where these boundaries lie can impact the course of treatment, which can include surgery or radiotherapy. The researches describe the boundary detection process as a “multi-parametric multi-class learning problem.”
Each participating site prepares its own data with a harmonizing process that can account for different methodologies between locations. The mpMRI scans are converted from the Digital Imaging and Communication in Medicine (DICOM) format to the Neuroimaging Informatics Technology Initiative (NIfTI) format which also eliminates any patient-identifiable information in the process. After some additional processing which maps and normalizes the scans as voxels, a mask is applied to remove all non-brain tissue from the images. The mpMRI scans are randomly sampled via [128, 128, 128]-sized patches, which the authors note is able to fit within the memory of a discrete GPU with at least 11GB of VRAM.
Once the data is collected by each collaborator, the training is conducted by passing a public initial model to the site. Each site trained against their own data for an epoch, then shared updates with the central server. The results were averaged according to the weight of each collaborator’s data, and then the process repeated in what are called federated rounds.
This is not the first FL application in healthcare, but is the largest of its kind to date. FL has previously been studied against CL and shown to have “almost identical performance.” It has also been used to assess breast cancer classification across 5 sites and to predict future oxygen requirements for COVID-19 patients using x-ray images and medical records across 20 sites. This study also represents the largest and most diverse dataset of glioblastoma patients ever studied, regardless of machine learning application.
The authors say this study’s success can enable further healthcare studies of rare diseases and underserved populations, facilitate additional understanding of glioblastoma using their consensus model, and improve multi-site collaborations with less direct data-sharing. You can read the full research article in Nature.
Top Photo by Tima Miroshnichenko